Scripting module
Script your own high-speed measurement sequences on the real-time system with scripting module SI 5
When speed and precise timing matter, measurement routines just can’t be fast enough. With a time-deterministic approach and 50 μs time interval between commands, scripting significantly boosts execution speed and reduces measurement time.
The module is seamlessly integrated in standard measurement modules: Scripts can be easily called from other modules, and custom functions or pre-defined measurement can be started from within a script.
The scripting module is not intended as a replacement of the Nanonis Programming Interface, but as complementary module: It allows 100x faster execution speed while the Programming Interface offers more flexibility.
See below for some script examples.
Switching spectroscopy PFM script courtesy of I. Gaponenko, University of Geneva
KEY FEATURES
- Fast and precise: Up to 20’000 commands/s executed on the real-time system
- Seamless integration with standard modules and programming interface
- FOR-loops for repeated measurement loops
- IF-commands for conditional execution and real-time feedback
- Data acquisition options for customized measurements
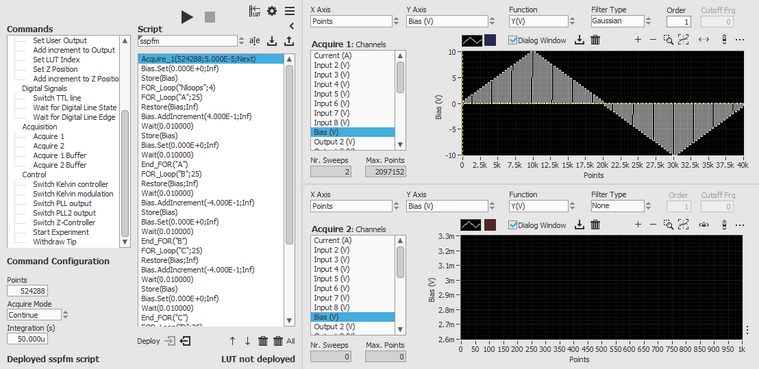
Script example 1: Switching spectroscopy PFM
The following script takes care of generating the switching bias pattern required for switching spectroscopy PFM measurements. The script consists of 4 main FOR loops, one for each quarter period of the waveform.
Note that these are not the input commands of the scripting module, this is the script file generated by the module (Script courtesy of I. Gaponenko, University of Geneva)
Acquire_1(524288;5.000E-5;Next)
Bias.Set(0.000E+0;Inf)
Store(Bias)
FOR_Loop("Nloops";4)
FOR_Loop("A";25)
Restore(Bias;Inf)
Bias.AddIncrement(4.000E-1;Inf)
Wait(0.010000)
Store(Bias)
Bias.Set(0.000E+0;Inf)
Wait(0.010000)
End_FOR("A")
FOR_Loop("B";25)
Restore(Bias;Inf)
Wait(0.010000)
Bias.AddIncrement(-4.000E-1;Inf)
Store(Bias)
Bias.Set(0.000E+0;Inf)
Wait(0.010000)
End_FOR("B")
FOR_Loop("C";25)
Restore(Bias;Inf)
Bias.AddIncrement(-4.000E-1;Inf)
Wait(0.010000)
Store(Bias)
Bias.Set(0.000E+0;Inf)
Wait(0.010000)
End_FOR("C")
FOR_Loop("D";25)
Restore(Bias;Inf)
Wait(0.010000)
Bias.AddIncrement(+4.000E-1;Inf)
Store(Bias)
Bias.Set(0.000E+0;Inf)
Wait(0.010000)
End_FOR("D")
End_FOR("Nloops")
Acquire_1_Buffer(SweepN=SweepN+1)
[Acquisition 1]
Channels=16777215
Channel X=-1
Channel Y=8
Display vs V=0
Filter type=0
Order=1
Cutoff Frq=0.125000
[Acquisition 2]
Channels=1
Channel X=0
Channel Y=0
Display vs V=0
Filter type=0
Order=1
Cutoff Frq=0.125000
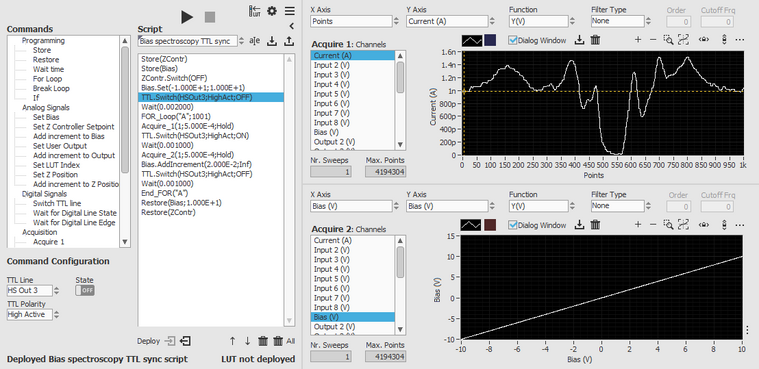
Script example 2: Triggered bias spectroscopy
This script runs a spectroscopy measurement with two data buffers: the bias is increased, data is acquired in a first buffer, then a TTL line is set to high and data is acquired into a second buffer. This script is used in combination with a laser and allows to record the response of a sample at each bias voltage setting before and after laser irradiation.
Note that these are not the input commands of the scripting module, this is the script file generated by the module.
Store(ZContr)
Store(Bias)
ZContr.Switch(OFF)
Bias.Set(-1.000E+0;1.000E+1)
TTL.Switch(HSOut3;HighAct;OFF)
Wait(0.002000)
FOR_Loop("A";1001)
Acquire_1(1;5.000E-4;Hold)
TTL.Switch(HSOut3;HighAct;ON)
Wait(0.001000)
Acquire_2(1;5.000E-4;Hold)
Bias.AddIncrement(2.000E-3;Inf)
TTL.Switch(HSOut3;HighAct;OFF)
Wait(0.001000)
End_FOR("A")
Restore(Bias;1.000E+1)
Restore(ZContr)